Your Genotyping Solution
The NEBNext Direct Genotyping Solution combines highly multiplexed, capture-based enrichment with maximum efficiency next generation sequencing to deliver cost-effective, high-throughput genotyping for a wide variety of applications.
Applicable for ranges spanning 100-5,000 markers, pre-capture multiplexing of up to 96 samples combined with 8 standard pool-indexes allows analysis of up to 768 samples / 3.8 million genotypes in a single Illumina sequencing run.
More pool-indexes are available upon request, allowing for multiplexing of up to 9,160 samples in a single run.
NEB will develop and balance bait sets using purified genomic DNA from your source material. This allows us to optimize performance for your specific application, with the quality of DNA that is typically obtained.
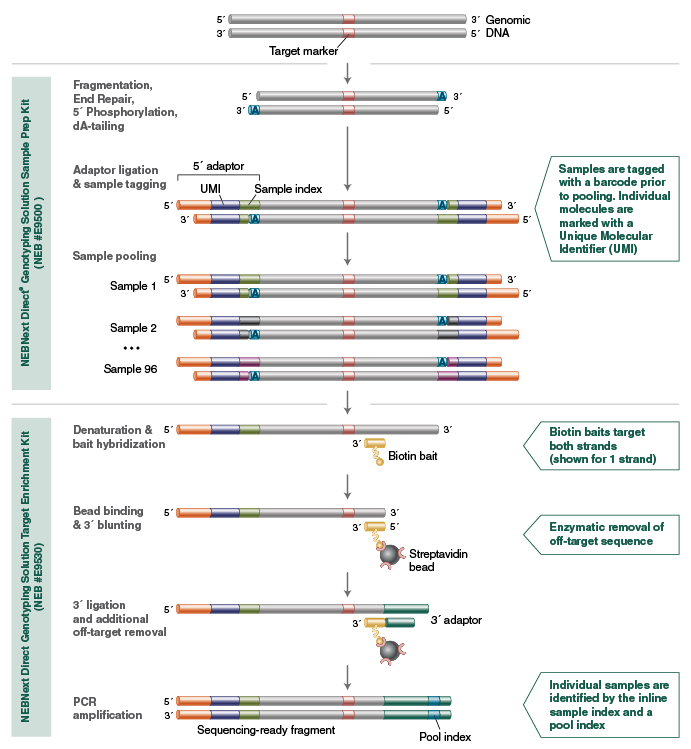
NEBNext Direct Genotyping Solution Workflow
The NEBNext Direct Genotyping Solution begins with 25-100 ng of purified genomic DNA. The DNA molecules are enzymatically fragmented and 5 ́ tagged with an Illumina-compatible P5 adaptors, incorporating both an inline sample index to tag each sample prior to pooling and an inline Unique Molecular Identifier (UMI) to mark each unique DNA fragment within the samples.
Up to 96 samples are subsequently pooled together prior to hybridization-based enrichment using biotinylated baits and captured on streptavidin beads. For the remainder of the protocol, up to 96 samples are processed as a single pool through ligation of a 3 ́ adaptor, removal of off-target sequence and final PCR, which amplifies the material and adds a second pool index to produce the final sequencing-ready fragment.
- Single-day workflow
- 96-plex pre-capture sample multiplexing of 100-5,000 markers
- Process up to 9,160 samples in a single sequencing run
- Bait design and sample multiplexing to maximize sequencer efficiency
- High specificity and coverage uniformity
Performance
Coverage Uniformity
The NEBNext Direct Genotyping Solution produces reliable and accurate genotyping calls using fewer sequencing reads. This is achieved through the combination of highly specific and highly consistent enrichment across targeted loci, resulting in less sequencing data due to off-target sequence, while ensuring more markers can be included in analyses.
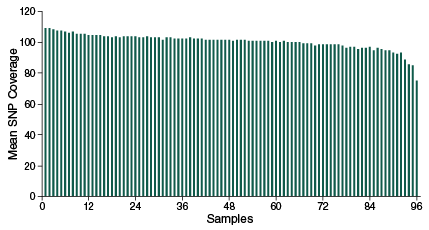
NEBNext Direct Genotyping Solution demonstrates similar coverage across 96 pooled samples
Mean SNP coverage of 2,309 SolCAP markers across 96 samples. 25 ng of purified tomato DNA was used for each sample. Samples were index-tagged and pooled prior to hybridization and Libraries were sequenced on an Illumina MiSeq with 20 cycles of Read 1 to sequence the 12 base UMI and 8 base sample index, and 75 cycles of Read 2 to sequence the targets.
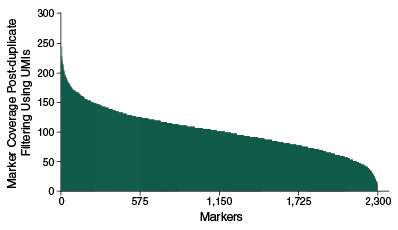
Mean Coverage across 2309 markers within a single sample
Histogram of coverage across each of the 2,309 SolCAP markers demonstrates evenness of enrichment across targets and coverage levels sufficient for genotyping calls. These data represent enrichment of a single tomato sample pooled with 95 others prior to hybridization. 25 ng of purified tomato DNA was used for each sample. Samples were index-tagged and pooled prior to hybridization and libraries were sequenced on an Illumina MiSeq with 20 cycles of Read 1 to sequence the 12 base UMI and 8 base sample index, and 75 cycles of Read 2 to sequence the targets.
Highly specific enrichment
Pre-capture pooling of 96 samples provides additional advantages, both in the significant reduction in pipetting steps, reducing plastic waste and consumables cost, as well as in the benefits from leveraging higher- throughput, lower cost sequencing platforms and configurations.
Up to 768 samples can be pooled together in a single sequencing run.
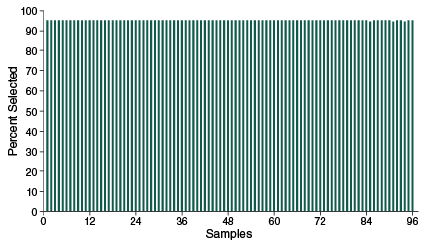
Specificity of enrichment of NEBNext Direct Genotyping Solution across 96 pooled samples
The percent of passing filter reads mapping to targeted regions demonstrates high specificity across 96 multiplexed samples using the NEBNext Direct Genotyping Solution. 25 ng of purified tomato DNA was used as input for each sample. Samples were index-tagged and pooled prior to hybridization and Libraries were sequenced on an Illumina MiSeq with 20 cycles of Read 1 to sequence the 12 base UMI and 8 base sample index, and 75 cycles of Read 2 to sequence the targets.
Further information can be found in our Technical Resources section or at neb.com




