Microbiome DNA analysis can be challenging due to the high percentage of host DNA present in many samples. The NEBNext Microbiome DNA Enrichment Kit facilitates enrichment of microbial genomic DNA from samples contain- ing methylated host DNA (including human), by selective binding and removal of the CpG-methylated host DNA. Importantly, microbial diversity remains intact after enrichment.
- Effective enrichment of microbial genomic DNA from samples contaminating host DNA
- Fast, simple protocol
- Enables microbiome whole genome sequencing, even for samples with high levels of host DNA
- Compatible with downstream applications including next generation sequencing on all platforms, qPCR and end point PCR
- Suitable for a wide range of sample types
- No requirement for live cells
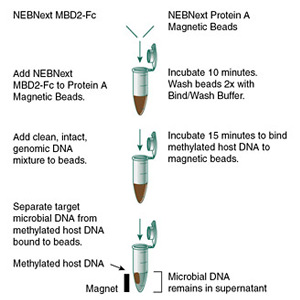
Workflow:
- Prepare Genomic DNA DNA should be free of proteins, proteinase A, SDS and organic solvents; size should be ≥ 15 kb for optimal performance.
- Combine MBD2-Fc and Magnetic Beads in 1X Bind/wash Buffer. Incubate the reaction for 10 minutes at room temperature. Wash beads two times in Bind/wash Reaction Buffer.
- Add DNA to MBD2-Fc Magnetic Beads. Incubate the reaction for 15 minutes at room temperature with gentle mixing.
- Collect Supernatant Fraction containing enriched microbial DNA and Bead Fraction-Containing Host DNA.
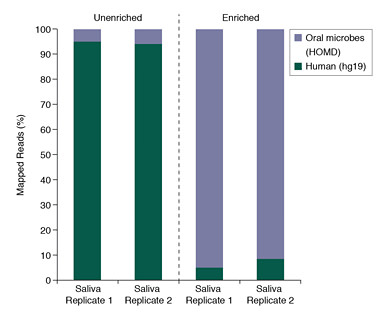
Salivary Microbiome DNA Enrichment
DNA was purified from pooled human saliva DNA (Innovative Research) and enriched using the NEBNext Microbiome DNA Enrichment Kit. Libraries were prepared from unenriched and enriched samples and sequenced on the SOLiD 4 platform. The graph shows percentages of 500M-537M SOLiD4 50bp reads that mapped to either the Human reference sequence (hg19) or to a microbe listed in Human Oral Microbiome Database (HOMD)[1]. (Because the HOMD collection is not comprehensive, ~80% of reads in the enriched samples do not map to either database.) Reads were mapped using Bowtie 0.12.7[2] with typical settings (2 mismatches in a 28 bp seed region, etc.).
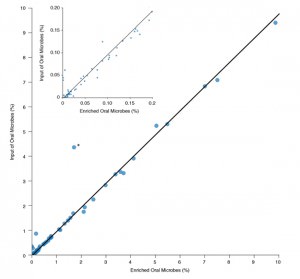
Microbiome Diversity
Microbiome Diversity is Retained after Enrichment with the NEBNext Microbiome DNA Enrichment Kit DNA was purified from pooled human saliva DNA (Innovative Research) and enriched using the NEBNext Microbiome DNA Enrichment Kit. Libraries were prepared from unenriched and enriched samples, followed by sequencing on the SOLiD 4 platform. The graph shows a comparison between relative abundance of each bacterial species listed in HOMD[1] before and after enrichment with the NEBNext Microbiome DNA Enrichment Kit. Abundance is inferred from the number of reads mapping to each species as a percentage of all reads mapping to HOMD. High concordance continues even to very low abundance species (inset). We compared 501M 50bp SOliD4 reads in the enriched dataset to 537M 50bp SOLiD4 reads in the unenriched dataset. Reads were mapped using Bowtie 0.12.7[2] with typical settings (2 mismatches in a 28bp seed region, etc). * Niesseria flavescens – This organism may have unusual methylation density, allowing it to bind the enriching beads at a low level. Other Niesseria species (N. mucosa, N. sicca and N. elognata) are represented, but do not exhibit this anomalous enrichment.
Further information can be found in our Technical Resources section or at neb.com