A high-performance alternative to bisulfite sequencing for methylome analysis
Whole Genome Bisulfite Sequencing (WGBS) has long been the gold standard for methylome analysis, but the chemical bisulfite reaction damages and degrades DNA, resulting in fragmentation and loss. Additionally, bisulfite libraries demonstrate significant GC bias and are enriched for methylated regions.
To overcome these limitations, we developed an enzyme-based approach, NEBNext Enzymatic Methyl-seq (EM-seq), a new method for identification of 5mC and 5hmC.
- Superior sensitivity of detection of 5-mC and 5-hmC
- Greater mapping efficiency
- More uniform GC coverage
- Detect more CpGs with fewer sequence reads
- Uniform dinucleotide distribution
- Larger library insert sizes
- High-efficiency library preparation
- Conversion module also avaible separately
The highly effective enzymatic conversion in this method minimizes damage to DNA and, with the supplied NEBNext UltraTM II library preparation workflow reagents, produces high quality libraries that enable superior detection of 5mC and 5hmC from fewer sequencing reads. Conveniently, the EM-seq method results in the same converted sequence as WGBS and so the same analysis pipelines can be used.
Video: EM-seq Workflow
Want to try the new NEBNext Enzymatic Methyl-seq? Request your sample today:
Feedback from our « early access » users:
|
„Em-Seq (…) enables us to determine in a precise and DNA sparing way the cytosine methylation status even at low integrity DNA. (…) It also opens new avenues to explorations of methylation at intact long DNA fragments.“ Vladimir Benes, Head Genomics Core Facility at EMBL Heidelberg |
||||
 |
||||
|
„Enzymatic conversion of EM-Seq is THE alternative and our comparisons clearly showed, that the quality of data obtained is better than with conventional bisulfite conversion.“ Dr. Alexander Vogt, Sequencing Specialist, Vienna BioCenterCore Facilities |
||||
 |
||||
There’s a new alternative to bisulfite sequencing
The NEBNext EM-seq method can be adopted for different sequencing platforms:
Illumina Plateforms:
The NEBNext Enzymatic Methyl-seq Kit (#E7120) contains all reagents needed for enzymatic conversion of methylated DNA and library preparation for Illumina sequencing.
Other plateforms:
The NEBNext Enzymatic Methyl-seq Conversion Module (#E7125) can be used for enzymatic DNA conversion for subsequent library prep.
Workflow NEBNext Enzymatic Methyl-seq Kit (#E7120) for Illumina:
Libraries are prepared using as little as 10 ng input DNA and the supplied NEBNext Ultra II reagents and the optimized EM-seq Adaptor. TET2 then oxidizes 5-mC and 5-hmC, providing protection from deamination by APOBEC in the next step. In contrast, unmodified cytosines are deaminated to uracils. Libraries are then amplified using a NEBNext master mix formulation of Q5U (a modified version of Q5 High-Fidelity DNA Polymerase), and sequenced using Illumina instrumentation.
The consistently high conversion performance and minimized DNA damage with the EM-seq protocol, in combination with highly efficient Ultra II library prep, result in superior detection of CpGs with fewer sequencing reads.
Multiplexing made easy:
The NEBNext Enzymatic Methyl-seq Kit (#E7120) contains Unique Dual Index Primer Pairs for 24-plex. The 96-well NEBNext Multiplex Oligos for Enzymatic Methyl-seq (Unique Dual Index Primer Pairs) (#E7140L) can be used in addition to perform up to 120-plex assays.
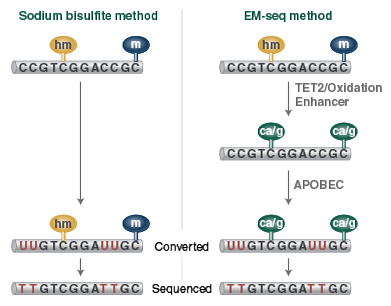
Sodium bisulfite treatment of DNA results in the deamination of cytosines to uracils, while the modified forms of cytosine (5mC and 5hmC) are not deaminated. When bisulfite treated DNA is PCR amplified, uracils are replaced by thymines, and 5mC and 5hmC are replaced by cytosines. Once sequenced, unmethylated cytosines are represented by thymines and 5mC and 5hmC are represented by cytosines. By comparing sequences to reference sequences (C/T and G/A converted genome), the methylation status can be assessed.
The first EM-seq conversion step uses TET2 and an Oxidation Enhancer to protect modified cytosines from downstream deamination. TET2 enzymatically oxidizes 5mC and 5hmC through a cascade reaction into 5-carboxycytosine. This protects 5mC and 5hmC from deamination. 5hmC can also be protected from deamination by glucosylation to form 5ghmc using the Oxidation Enhancer. The second enzymatic step uses APOBEC, which deaminates cytosine but does not affect 5caC and 5ghmC.The resulting converted sequence is the same as that for bisulfite-treated DNA and so can be analyzed in the same way.
Advantages of the NEBNext Enzymatic Methyl-seq method
Larger library insert sizes
The more gentle treatment of DNA by the steps in the EM-seq workflow, compared to the harsh bisulfite treatment, minimizes damage to DNA. As a result, EM-seq converted DNA is more intact than bisulfite converted DNA, resulting in libraries with a higher percentage of longer inserts, as shown below. This enables longer sequencing reads, resulting in greater confidence in mapping, and potentially lower per-base sequencing costs depending on instrumentation and other sequencing reaction details.
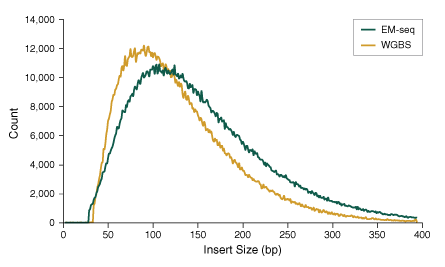
50 ng Human NA12878 genomic DNA was sheared to 300 bp using the Covaris® S2 instrument and used as input into EM-seq and WGBS protocols. For WGBS, NEBNext Ultra II DNA was used for library construction, followed by the Zymo Research EZ DNA Methylation-GoldTM kit for bisulfite conversion. Libraries were sequenced on an Illumina® MiSeq® (2 x 76 bases) and insert sizes were determined using Picard 2.18.14. The normalized frequency of each insert size was plotted, illustrating that library insert sizes are larger for EM-seq than for WGBS, and indicating that EM-seq does not damage DNA as bisulfite treatment does in WGBS.
Increased library yields
The DNA damage, fragmentation and loss that result from bisulfite treatment reduce yields of bisulfite-converted libraries after amplification. In contrast, the more gentle treatment in the EM-seq workflow allows maintenance of high quality DNA libraries, which can be efficiently amplified. As a result, EM-seq library yields are not only higher than WGBS library yields, but are achieved with fewer PCR cycles.
Importantly, these higher yields are not due to the presence of PCR duplicates, which can be especially apparent at low input amounts. Indeed, EM-seq libraries display consistently low levels of duplicates across a range of input amounts.
Uniformity of GC coverage
While sufficient yield of a library is required for successful sequencing, the quality of a library is also critical. A high-quality library will have uniform representation of the original sample, including uniform coverage across the GC spectrum.
Since bisulfite treatment acts upon and damages unmethylated cytosines, which comprise the majority of cytosines, this harsh treatment therefore disproportionately affects GC-containing regions. This context-specific damage, breakage and loss lead to bisulfite- treated libraries being under-represented for GC content and over-represented for AT. In contrast, EM-seq libraries show uniform GC coverage, highlighting the lack of damage to DNA, and that the libraries are representative of the original sample.
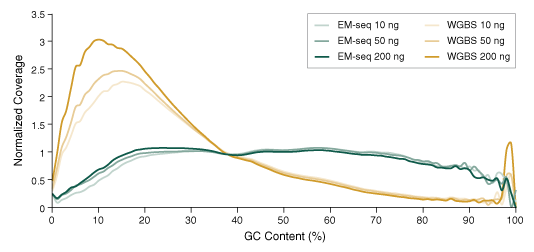
10, 50 and 200 ng Human NA12878 genomic DNA was sheared to 300 bp using the Covaris S2 instrument and used as input into EM-seq and WGBS protocols. For WGBS, NEBNext Ultra II DNA was used for library construction, followed by the Zymo Research EZ DNA Methylation-Gold Kit for bisulfite conversion. Libraries were sequenced on an Illumina NovaSeq® 6000 (2 x 100 bases). Reads were aligned to hg38 using bwa-meth 0.2.2. GC coverage was analyzed using Picard 2.18.14 and the distribution of normalized coverage across different GC contents of the genome (0-100%) was plotted. EM-seq libraries have significantly more uniform GC coverage, and lack the AT over-representation and GC under-representation typical of WGBS libraries.
View or download extensive performance data in our Technical Note.
NEBNext Enzymatic Methyl-seq products available:
As of: 07.05.2019
NEBNext products are available at a permanent low price that can not be further discounted.
Further information can be found in our Technical Resources section or at neb.com


